Note
Go to the end to download the full example code.
How to use SKADA
This is a short example to get started with SKADA and perform domain adaptation on a simple dataset. It illustrates the API choice specific to DA.
# Author: Remi Flamary
#
# License: BSD 3-Clause
# sphinx_gallery_thumbnail_number = 1
import matplotlib.pyplot as plt
import numpy as np
from sklearn.decomposition import PCA
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import GridSearchCV, cross_val_score
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import StandardScaler
from sklearn.svm import SVC
from skada import (
CORAL,
CORALAdapter,
GaussianReweightAdapter,
PerDomain,
SelectSource,
SelectSourceTarget,
make_da_pipeline,
source_target_split,
)
from skada.datasets import make_shifted_datasets
from skada.metrics import PredictionEntropyScorer
from skada.model_selection import SourceTargetShuffleSplit
DA dataset
We generate a simple 2D DA dataset. Note that DA datasets provided by SKADA are organized as follows:
Xis the input data, including the source and the target samplesyis the output data to be predicted (labels on target samples are not used when fitting the DA estimator)sample_domainencodes the domain of each sample (integer >=0 for source and <0 for target)
# Get DA dataset
X, y, sample_domain = make_shifted_datasets(
20, 20, shift="conditional_shift", random_state=42
)
# split source and target for visualization
Xs, Xt, ys, yt = source_target_split(X, y, sample_domain=sample_domain)
sample_domain_s = np.ones(Xs.shape[0])
sample_domain_t = -np.ones(Xt.shape[0]) * 2
# plot data
plt.figure(1, (10, 5))
plt.subplot(1, 2, 1)
plt.scatter(Xs[:, 0], Xs[:, 1], c=ys, cmap="tab10", vmax=9, label="Source")
plt.title("Source data")
ax = plt.axis()
plt.subplot(1, 2, 2)
plt.scatter(Xt[:, 0], Xt[:, 1], c=yt, cmap="tab10", vmax=9, label="Target")
plt.axis(ax)
plt.title("Target data")
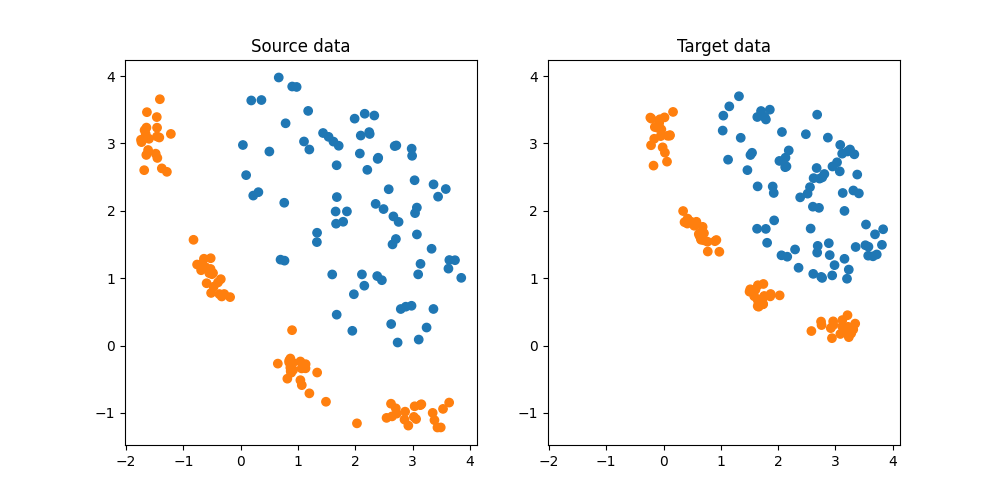
Text(0.5, 1.0, 'Target data')
DA Classifier estimator
SKADA estimators are used like scikit-learn estimators. The only difference is
that the sample_domain array must be passed by name when fitting the
estimator.
# create a DA estimator
clf = CORAL()
# train on all data
clf.fit(X, y, sample_domain=sample_domain)
# estimator is designed to predict on target by default
yt_pred = clf.predict(Xt)
# accuracy on source and target
print("Accuracy on source:", clf.score(Xs, ys))
print("Accuracy on target:", clf.score(Xt, yt))
Accuracy on source: 0.84375
Accuracy on target: 1.0
DA estimator in a pipeline
# SKADA estimators can be used as the final estimator of a scikit-learn pipeline.
# Again, the only difference is that the :code:`sample_domain` array must be passed
# by name during in fit.
# create a DA pipeline
pipe = make_pipeline(StandardScaler(), CORAL(base_estimator=SVC()))
pipe.fit(X, y, sample_domain=sample_domain)
print("Accuracy on target:", pipe.score(Xt, yt))
Accuracy on target: 1.0
DA Adapter pipeline
Several SKADA estimators include a data adapter that transforms the input data
so that a scikit-learn estimator can be used. For those methods, SKADA
provides a Adapter class that can be used in a DA pipeline from
make_da_pipeline.
Here is an example with the CORAL and GaussianReweight adapters.
# Note that as illustrated below for reweighting adapters, one needs a
# subsequent estimator that takes :code:`sample_weight` as an input parameter.
# This can be done using the :code:`set_fit_request` method of the estimator
# by calling :code:`.set_fit_request(sample_weight=True)`.
# If the estimator (for pipeline or DA estimator) does not
# require sample weights, the DA pipeline will raise an error.
# create a DA pipeline with CORAL adapter
pipe = make_da_pipeline(StandardScaler(), CORALAdapter(), SVC())
pipe.fit(X, y, sample_domain=sample_domain)
print("Accuracy on target:", pipe.score(Xt, yt))
# create a DA pipeline with GaussianReweight adapter (does not work well on
# conditional shift).
pipe = make_da_pipeline(
StandardScaler(),
GaussianReweightAdapter(),
LogisticRegression().set_fit_request(sample_weight=True),
)
pipe.fit(X, y, sample_domain=sample_domain)
print("Accuracy on target:", pipe.score(Xt, yt))
Accuracy on target: 1.0
Accuracy on target: 0.5
DA estimators with score cross-validation
DA estimators are compatible with scikit-learn cross-validation functions.
Note that the sample_domain array must be passed in the params
dictionary of the cross_val_score function.
# splitter for cross-validation of score
cv = SourceTargetShuffleSplit(random_state=0)
# DA scorer not using target labels (not available in DA)
scorer = PredictionEntropyScorer()
clf = CORAL(SVC(probability=True)) # needs probability for entropy score
# cross-validation
scores = cross_val_score(
clf, X, y, params={"sample_domain": sample_domain}, cv=cv, scoring=scorer
)
print(f"Entropy score: {scores.mean():1.2f} (+-{scores.std():1.2f})")
Entropy score: -0.04 (+-0.01)
DA estimator with grid search
DA estimators are also compatible with scikit-learn grid search functions.
Note that the sample_domain array must be passed in the fit
method of the grid search.
reg_coral = [0.1, 0.5, 1, "auto"]
clf = make_da_pipeline(StandardScaler(), CORALAdapter(), SVC(probability=True))
# grid search
grid_search = GridSearchCV(
estimator=clf,
param_grid={"coraladapter__reg": reg_coral},
cv=SourceTargetShuffleSplit(random_state=0),
scoring=PredictionEntropyScorer(),
)
grid_search.fit(X, y, sample_domain=sample_domain)
print("Best regularization parameter:", grid_search.best_params_["coraladapter__reg"])
print("Accuracy on target:", np.mean(grid_search.predict(Xt) == yt))
Best regularization parameter: auto
Accuracy on target: 1.0
Advanced DA pipeline
The DA pipeline can be used with any estimator and any adapter. But more importantly all estimators in the pipeline are automatically wrapped in what we call in skada a Selector. The selector is a wrapper that allows you to choose which data is passed during fit and predict/transform.
In the following example, one StandardScaler is trained per domain. Then a single SVC is trained on source data only. When predicting on target data the pipeline will automatically use the StandardScaler trained on target and the SVC trained on source.
# create a DA pipeline with SelectSourceTarget estimators
pipe = make_da_pipeline(
SelectSourceTarget(StandardScaler()),
SelectSource(SVC()),
)
pipe.fit(X, y, sample_domain=sample_domain)
print("Accuracy on source:", pipe.score(Xs, ys, sample_domain=sample_domain_s))
print("Accuracy on target:", pipe.score(Xt, yt)) # target by default
Accuracy on source: 1.0
Accuracy on target: 1.0
Similarly one can use the PerDomain selector to train a different estimator
per domain. This allows to handle multiple source and target domains. In this
case sample_domain must be provided to fit and predict/transform.
pipe = make_da_pipeline(
PerDomain(StandardScaler()),
SelectSource(SVC()),
)
pipe.fit(X, y, sample_domain=sample_domain)
print("Accuracy on all data:", pipe.score(X, y, sample_domain=sample_domain))
Accuracy on all data: 1.0
One can use a default selector on the whole pipeline which allows for instance to train the whole pipeline only on the source data as follows:
pipe_train_on_source = make_da_pipeline(
StandardScaler(),
SVC(),
default_selector=SelectSource,
)
pipe_train_on_source.fit(X, y, sample_domain=sample_domain)
print("Accuracy on source:", pipe_train_on_source.score(Xs, ys))
print("Accuracy on target:", pipe_train_on_source.score(Xt, yt))
Accuracy on source: 1.0
Accuracy on target: 0.5
One can also use a default selector on the whole pipeline but overwrite it for
the last estimator. In the example below a StandardScaler and a
PCA are estimated per domain but the final SVC is trained on source data only.
pipe_perdomain = make_da_pipeline(
StandardScaler(),
PCA(n_components=2),
SelectSource(SVC()),
default_selector=SelectSourceTarget,
mask_target_labels=False,
)
pipe_perdomain.fit(X, y, sample_domain=sample_domain)
print(
"Accuracy on source:", pipe_perdomain.score(Xs, ys, sample_domain=sample_domain_s)
)
print(
"Accuracy on target:", pipe_perdomain.score(Xt, yt, sample_domain=sample_domain_t)
)
Accuracy on source: 1.0
Accuracy on target: 1.0
Total running time of the script: (0 minutes 2.081 seconds)