Note
Go to the end to download the full example code.
Using cross_val_score with skada
This example illustrates the use of DA scorer such as TargetAccuracyScorer
with cross_val_score.
We first create a shifted dataset. Then we prepare the pipeline including a
base estimator doing the classification and the DA estimator. We use
ShuffleSplit as cross-validation strategy.
import matplotlib.pyplot as plt
import numpy as np
from sklearn.model_selection import ShuffleSplit, cross_val_score
from sklearn.svm import SVC
from skada import EntropicOTMapping, make_da_pipeline, source_target_split
from skada.datasets import make_shifted_datasets
from skada.metrics import SupervisedScorer
RANDOM_SEED = 0
dataset = make_shifted_datasets(
n_samples_source=30,
n_samples_target=20,
shift="conditional_shift",
label="binary",
noise=0.4,
random_state=RANDOM_SEED,
return_dataset=True,
)
base_estimator = SVC()
estimator = EntropicOTMapping(base_estimator=base_estimator, reg_e=0.5, tol=1e-3)
X, y, sample_domain = dataset.pack(
as_sources=["s"], as_targets=["t"], mask_target_labels=True
)
X_source, X_target, y_source, y_target = source_target_split(
X, y, sample_domain=sample_domain
)
cv = ShuffleSplit(n_splits=5, test_size=0.3, random_state=RANDOM_SEED)
The DA estimator pipeline is ready to be used with cross_val_score.
Source data from the training splits is first adapted with the target
data from the same splits and then used to fit the base estimator.
The target data from the test split is used to compute the score.
The separation between source and target data is done automatically
by the DA pipeline thanks to sample_domain. The target_labels
are only used by the SupervisedScorer.
_, target_labels, _ = dataset.pack(
as_sources=["s"], as_targets=["t"], mask_target_labels=False
)
scores_sup = cross_val_score(
estimator,
X,
y,
cv=cv,
params={"sample_domain": sample_domain, "target_labels": target_labels},
scoring=SupervisedScorer(),
)
print(
"Cross-validation score with supervised DA: "
f"{np.mean(scores_sup):.2f} (+/- {np.std(scores_sup):.2f})"
)
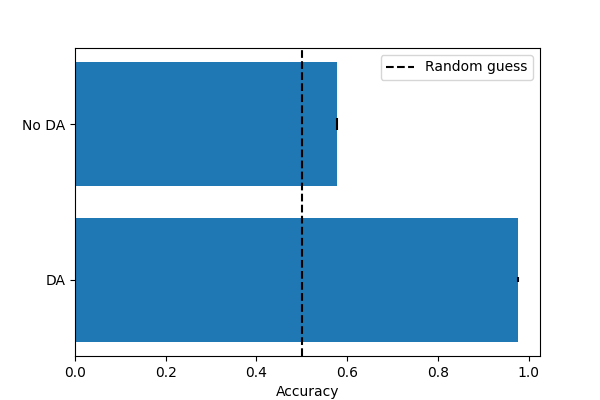
Cross-validation score with supervised DA: 0.98 (+/- 0.01)
To evaluate the performance of the DA estimator, we compare it with the
performance of the base estimator without DA. We use the same cross-validation
strategy and the same data splits. We create a DA pipeline with
make_da_pipeline including the base estimator only. The
sample_domain and target_labels are also passed to the pipeline
to separate the source and target data and to compute the score.
estimator_no_da = make_da_pipeline(base_estimator)
scores_no_da = cross_val_score(
estimator_no_da,
X,
y,
cv=cv,
params={"sample_domain": sample_domain, "target_labels": target_labels},
scoring=SupervisedScorer(),
)
print(
"Cross-validation score without DA: "
f"{np.mean(scores_no_da):.2f} (+/- {np.std(scores_no_da):.2f})"
)
Cross-validation score without DA: 0.58 (+/- 0.04)
plt.figure(figsize=(6, 4))
plt.barh(
[0, 1],
[np.mean(scores_sup), np.mean(scores_no_da)],
yerr=[np.std(scores_sup), np.std(scores_no_da)],
)
plt.yticks([0, 1], ["DA", "No DA"])
plt.xlabel("Accuracy")
plt.axvline(0.5, color="k", linestyle="--", label="Random guess")
plt.legend()
plt.show()
Total running time of the script: (0 minutes 0.344 seconds)