skada.NearestNeighborReweight
- skada.NearestNeighborReweight(base_estimator=None, n_neighbors=1, weights='uniform', algorithm='auto', leaf_size=30, p=2, metric='minkowski', metric_params=None, n_jobs=None, laplace_smoothing=False)[source]
Density re-weighting pipeline adapter and estimator.
The last 7 parameters are the parameters from the KNN estimator that will be used to estimate the weights in the adapt method
See [24] for details.
- Parameters:
- n_neighborsint, default=1
Number of neighbors to use for the KNN
- base_estimatorsklearn estimator, default=None
estimator used for fitting and prediction
- laplace_smoothingbool, default=False, optional
True if we want to use laplace smoothing, and thus adding 1 to all our weights (to prevent some of them to be 0)
- weights{'uniform', 'distance'}, callable or None, default='uniform'
Weight function used in prediction. Possible values:
'uniform' : uniform weights. All points in each neighborhood are weighted equally.
'distance' : weight points by the inverse of their distance. in this case, closer neighbors of a query point will have a greater influence than neighbors which are further away.
[callable] : a user-defined function which accepts an array of distances, and returns an array of the same shape containing the weights.
Refer to the example entitled Nearest Neighbors Classification showing the impact of the weights parameter on the decision boundary.
- algorithm{'auto', 'ball_tree', 'kd_tree', 'brute'}, default='auto'
Algorithm used to compute the nearest neighbors:
'ball_tree' will use
BallTree'kd_tree' will use
KDTree'brute' will use a brute-force search.
'auto' will attempt to decide the most appropriate algorithm based on the values passed to
fit()method.
Note: fitting on sparse input will override the setting of this parameter, using brute force.
- leaf_sizeint, default=30
Leaf size passed to BallTree or KDTree. This can affect the speed of the construction and query, as well as the memory required to store the tree. The optimal value depends on the nature of the problem.
- pfloat, default=2
Power parameter for the Minkowski metric. When p = 1, this is equivalent to using manhattan_distance (l1), and euclidean_distance (l2) for p = 2. For arbitrary p, minkowski_distance (l_p) is used. This parameter is expected to be positive.
- metricstr or callable, default='minkowski'
Metric to use for distance computation. Default is "minkowski", which results in the standard Euclidean distance when p = 2. See the documentation of scipy.spatial.distance and the metrics listed in
distance_metricsfor valid metric values.If metric is "precomputed", X is assumed to be a distance matrix and must be square during fit. X may be a sparse graph, in which case only "nonzero" elements may be considered neighbors.
If metric is a callable function, it takes two arrays representing 1D vectors as inputs and must return one value indicating the distance between those vectors. This works for Scipy's metrics, but is less efficient than passing the metric name as a string.
- metric_paramsdict, default=None
Additional keyword arguments for the metric function.
- n_jobsint, default=None
The number of parallel jobs to run for neighbors search.
Nonemeans 1 unless in ajoblib.parallel_backendcontext.-1means using all processors. See Glossary for more details. Doesn't affectfit()method.
- Returns:
- pipelinesklearn pipeline
Pipeline containing the DensityReweight adapter and the base estimator.
References
[24]Nearest neighbor-based importance weighting. In 2012 IEEE International Workshop on Machine Learning for Signal Processing, pages 1–6. IEEE.
Examples using skada.NearestNeighborReweight
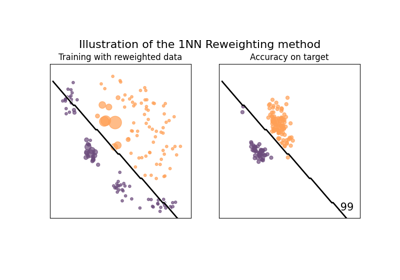
Reweighting method example on covariate shift dataset